OVITO Extensions Directory
A curated list of community extensions for OVITO Pro and the OVITO Python module
This page lists useful Python functions for OVITO, e.g., additional modifiers, file format readers, and viewport layers. These open-source extensions are contributed and maintained by independent members of the OVITO user community.
The extensions are hosted in external code repositories and can be easily installed in OVITO Pro or used with the standalone OVITO Python Module. Please see each extension's documentation or the manual for instructions.
How to submit your extension: If you would like your own development to be added to this list, please write a short email to the OVITO team at social@ovito.org. Thank you.
Modifiers
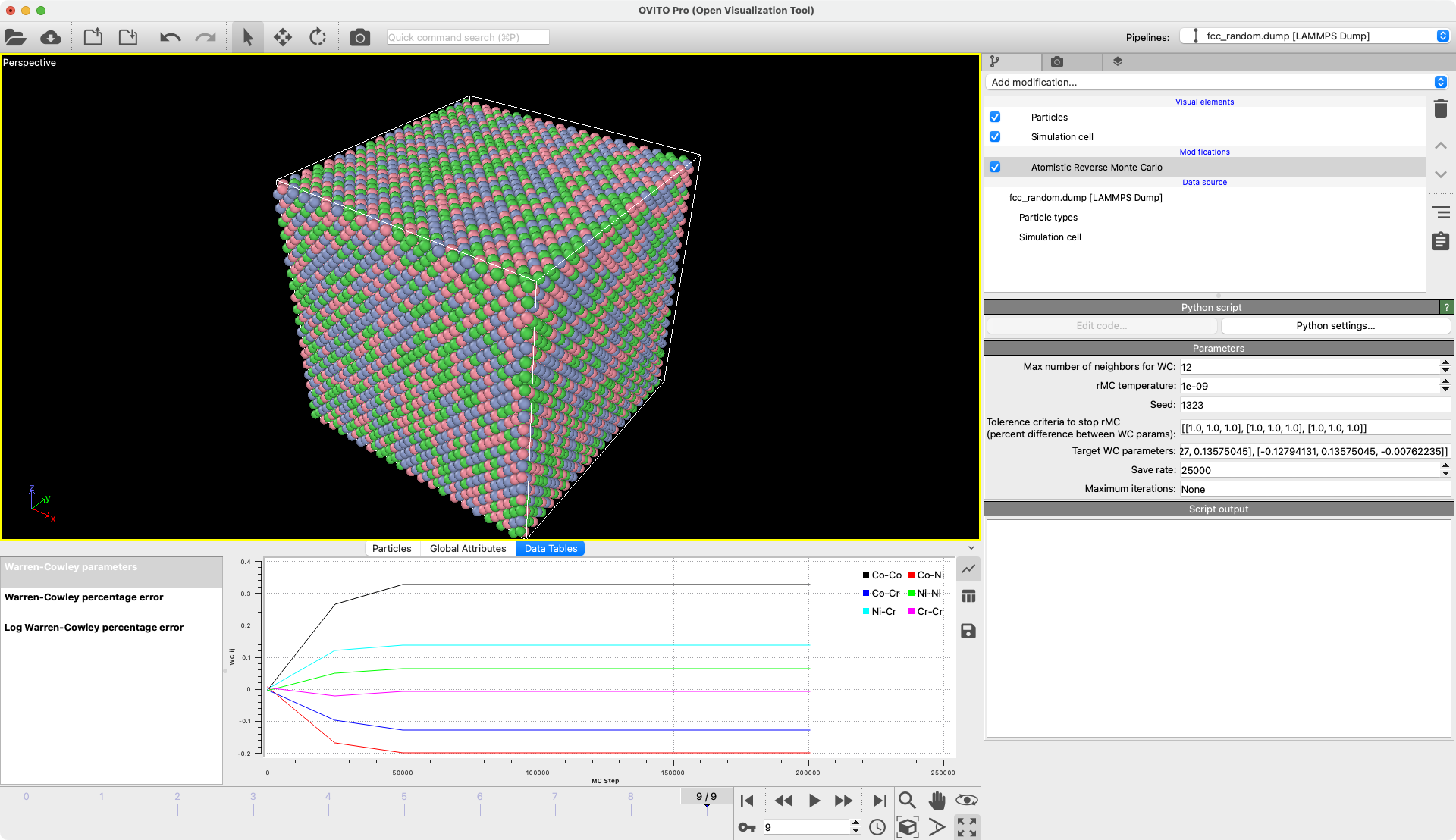
Atomistic Reverse Monte-Carlo
Generate bulk crystal structures with target Warren-Cowley parameters.
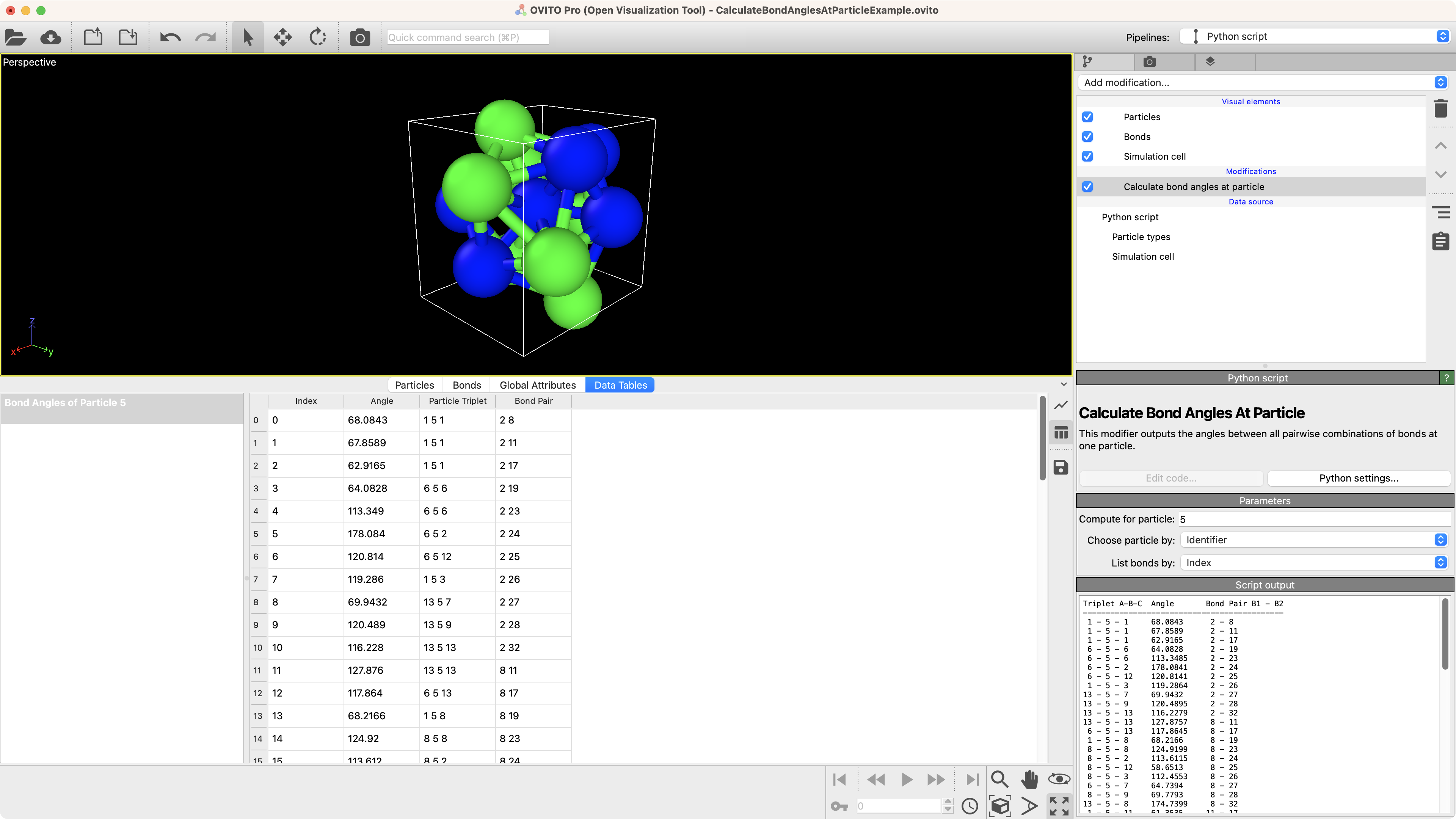
Calculate bond angles at particle
Compute the angles between all pairwise combinations of bonds of a selected particle.
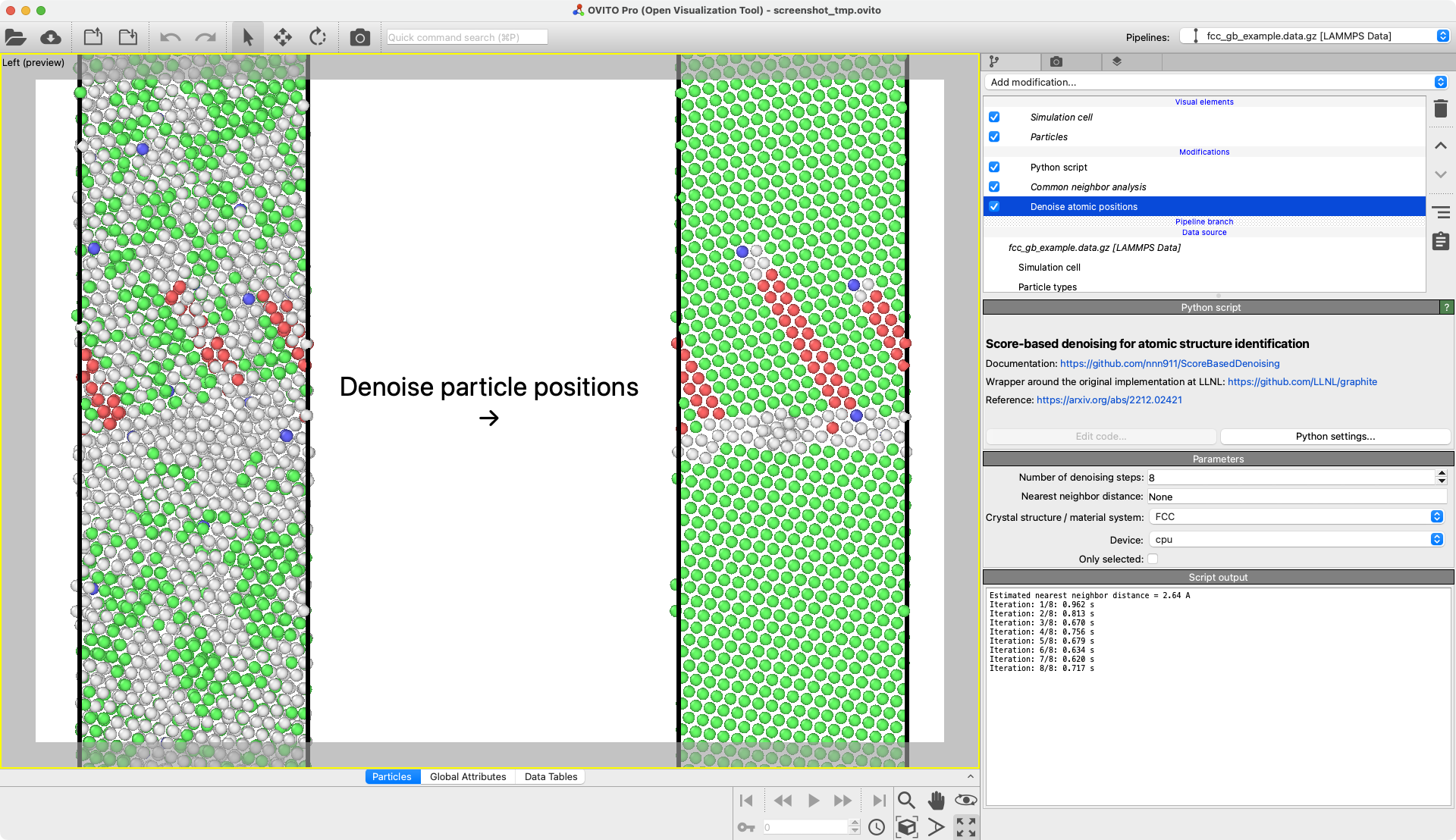
Denoise particle positions
Wrapper for the “Score-based denoising for atomic structure identification” method presented in this arXiv preprint and implemented here.
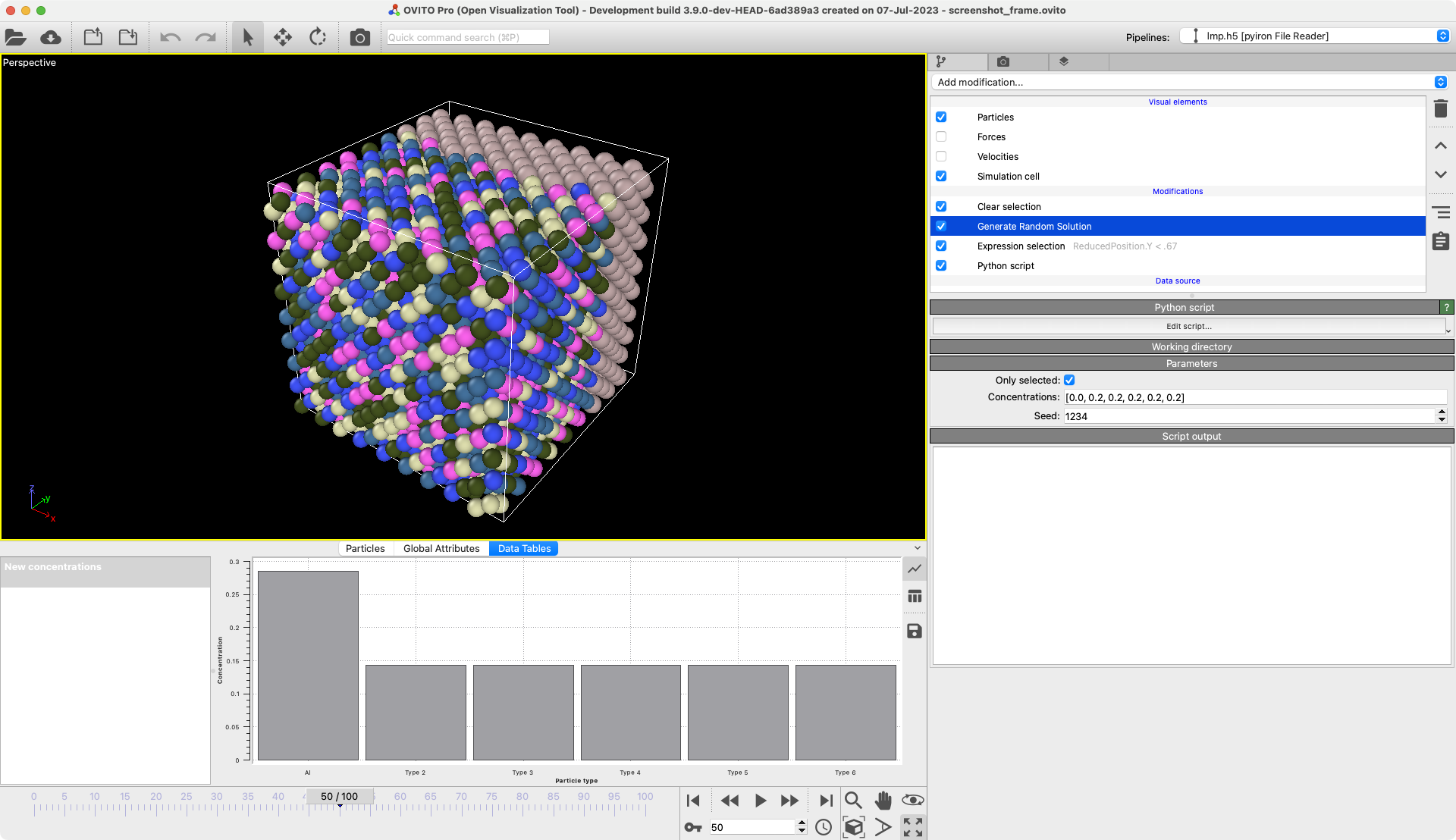
Generate random solution
Randomly change the types to particles to create a solution with given composition.
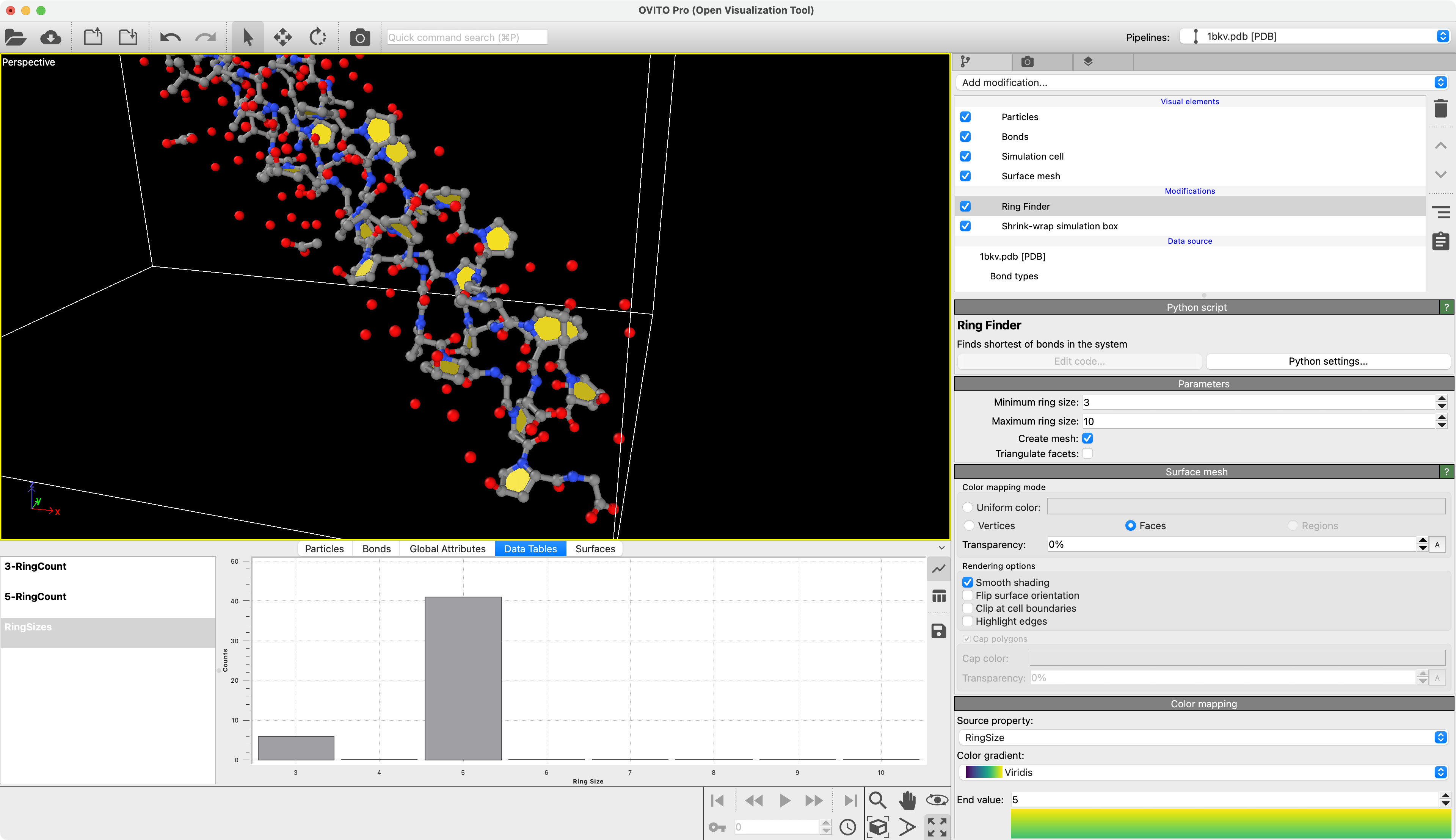
Ring Finder
Find shortest rings formed by bonds in a molecule. The rings are visualized as polygons.
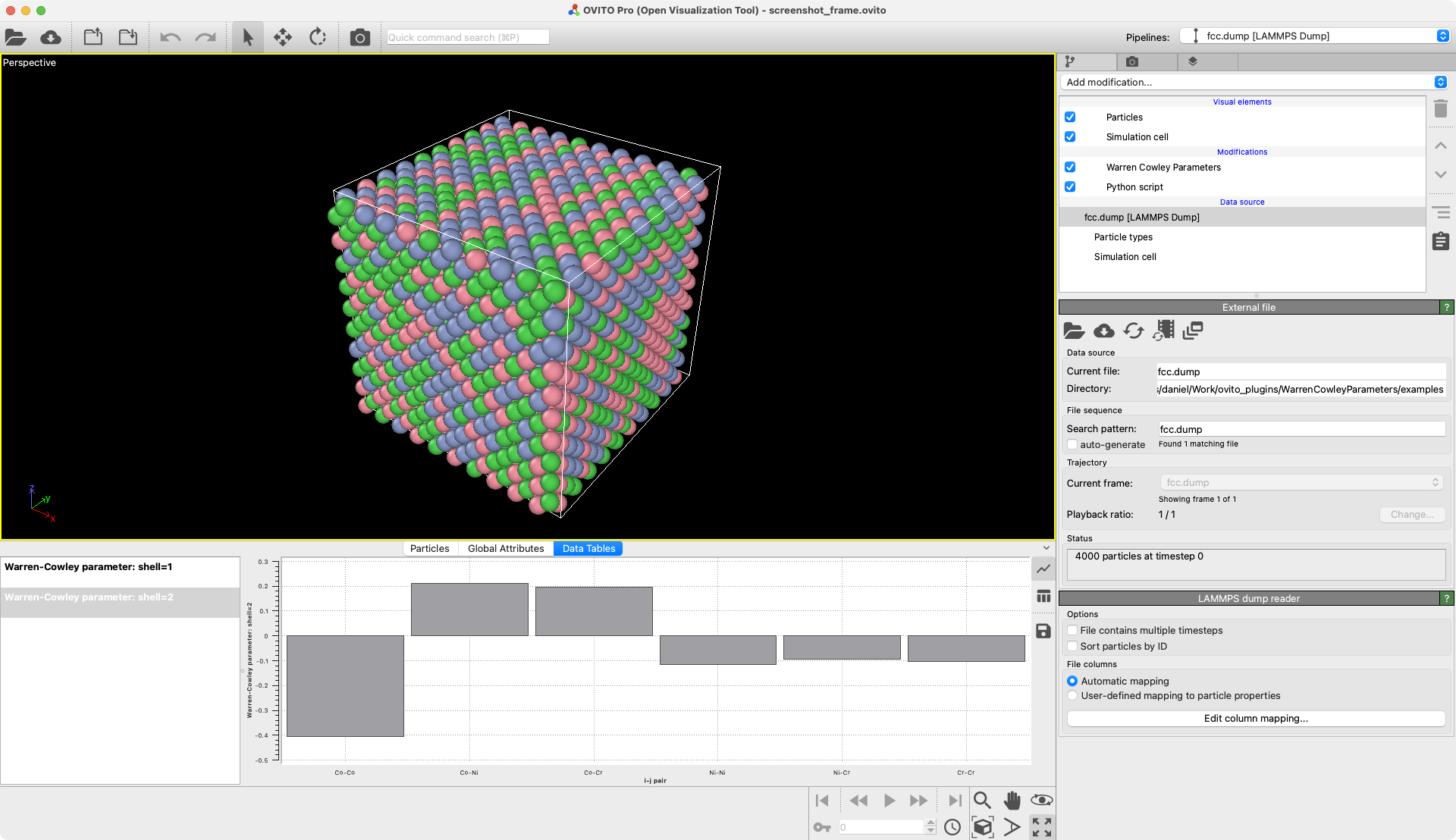
Warren-Cowley short-range order parameters
Calculate the chemical short-range order in an atomic system.
File Readers
Viewport Layers
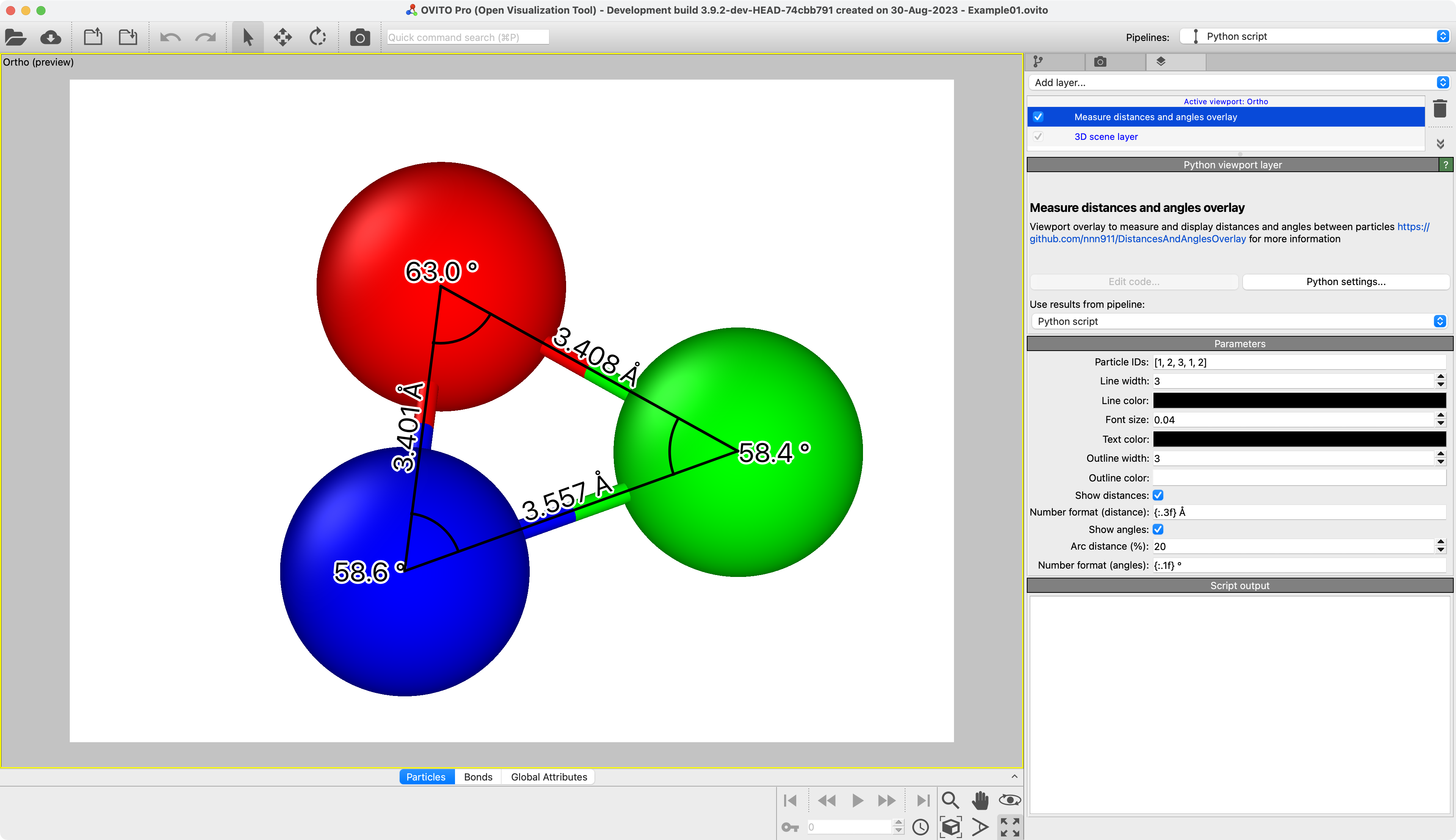
Measure Distances And Angles
Measure distances and angles between specific particles and visualize them.